Bailey Black, Microbiology & Molecular Genetics
Research: G-alpha 7 Phenotype Identification in Differentiation
[View my work!]
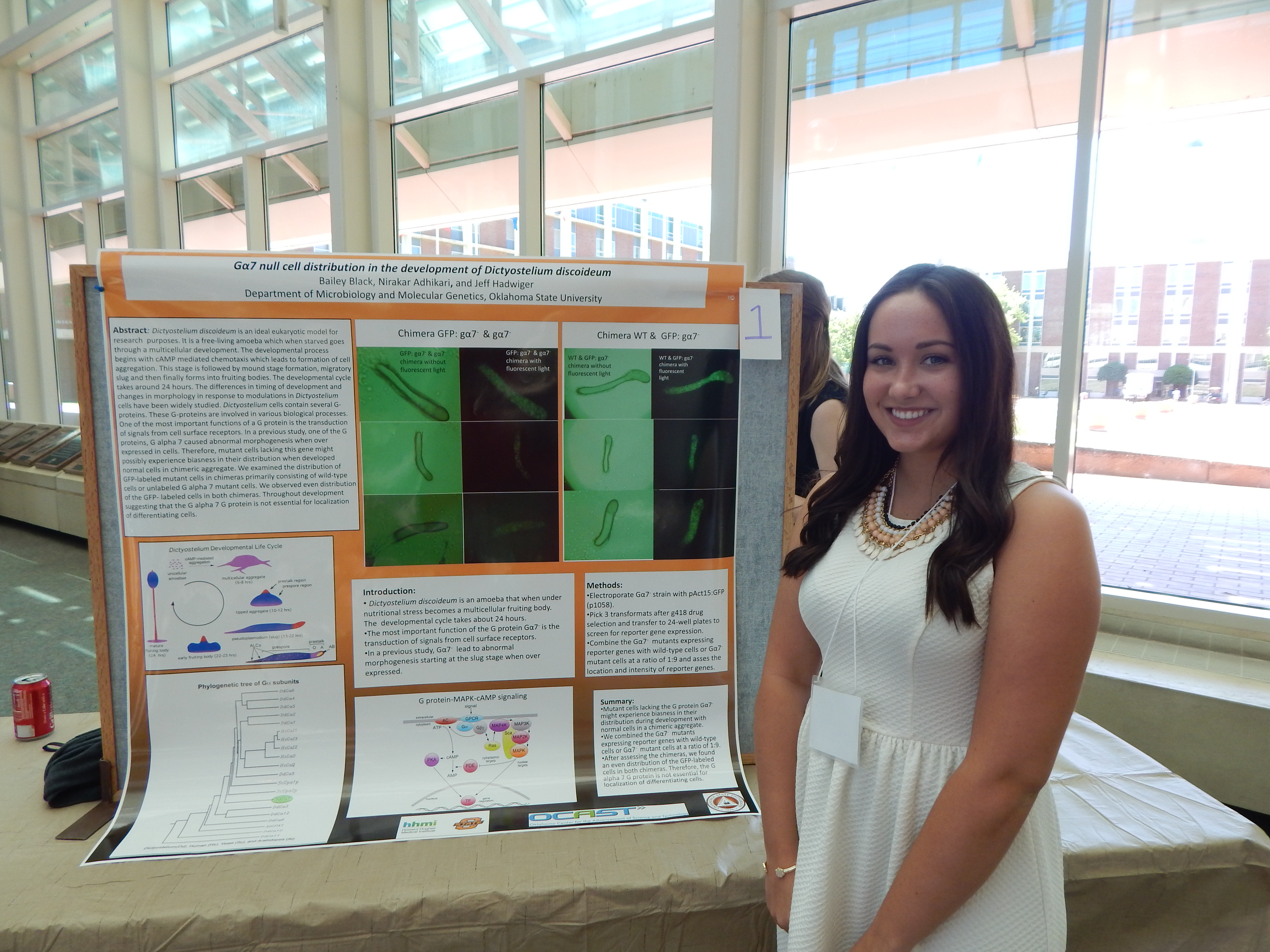
Project Summary: I will be looking at Dictyostelium cells during development and determining if a mutant strain containing G protein 7 has any effect on the development. When starved, Dictyostelium cells form into an aggegrate, then a slug,and then a fruiting body.
The G protein 7 will be dyed a green florescent color so it can be easily identified. To determine if there is an effect, I will form one chimera with the mutant strain containing the green fluorescent protein and the wild type cells. The other chimera will contain the mutant strain containing the green fluorescent protein and the mutant strain without the green fluorescent protein. I will be using a microscope to see if the green fluorescent protein shows up where it typically would during normal development. I will be looking at the slug form of the Dictyostelium cells. If the green fluorescent protein shows up where it normally does not then the G protein 7 must have an effect on the developmental cycle. From there, we would have to do further research to determine ifthe protein has an effect on the pre-spore cells, pre-stalk cells, or elsewhere in the developmental cycle.
In previous research some of the results found were premature sporulation, had no developmental defects, and cause spore deficiencies. I expect to see some of the same results. If the green fluorescent protein is found where it normally is then the G protein 7 has no effect on the developmental cycle. Iam also learning laboratory techniques such as how to conduct mini and large plasmid preparations, how to make medium, how to conduct an electroporation of Dictyostelium cells, and how to conduct an isolation of DNA from Agarose gels using Gene Clean 2. These techniques are all used in the project I am conducting.
Madison Faris, Communication Science & Disorders
Mentor: Dr.Sabiha Parveen, Communication Science & Disorders
Research: Executive Fuctioning in Parkinson Disease
[View my work!]
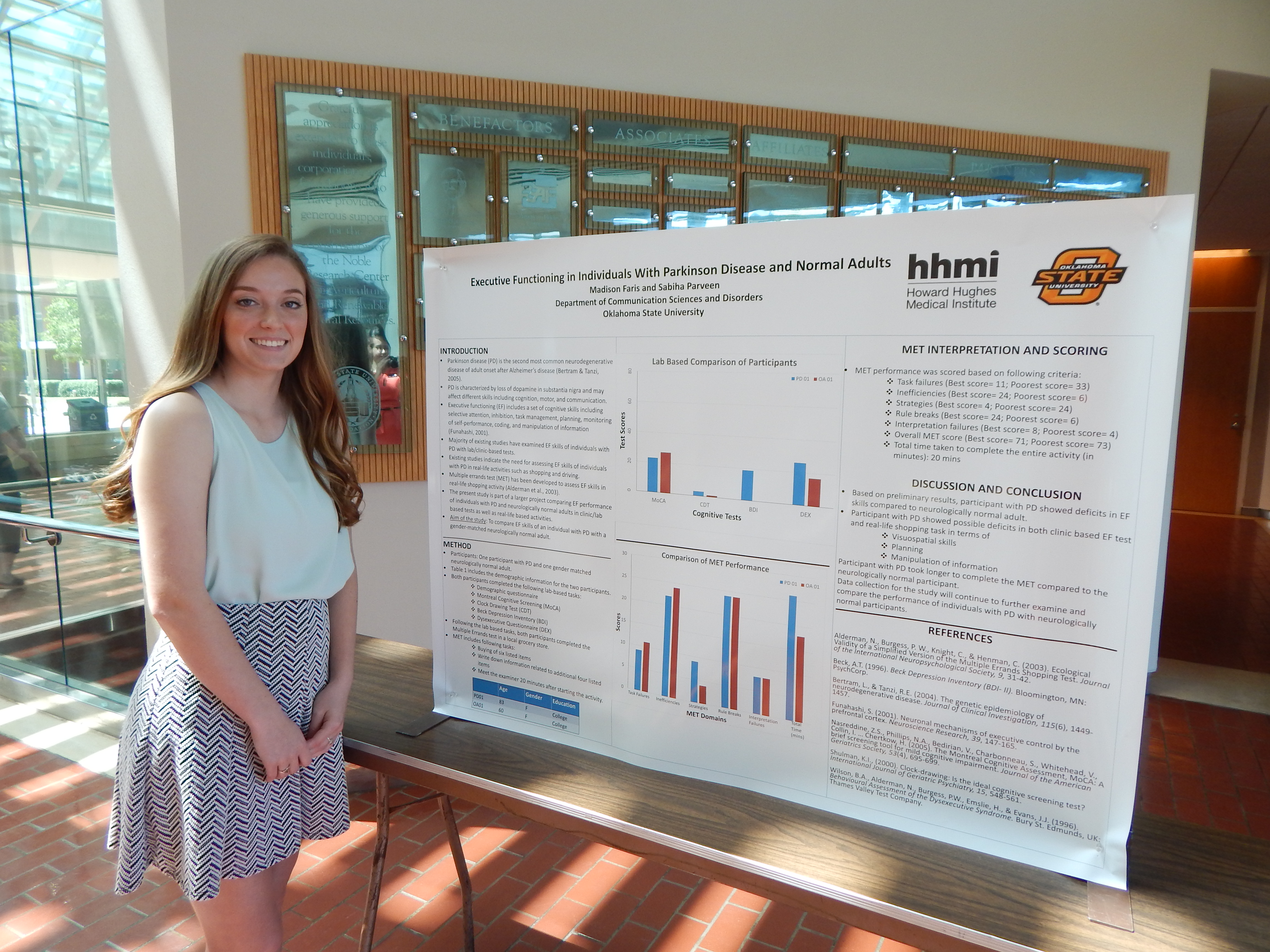
Project Summary: In Communication Sciences and Disorders, there are many areas that can be studied, as this is a broad range subject. Currently, I am researching the different levels of executive functioning in patients with Parkinson disease versus neurologically normal older and younger individuals. Executive functioning is the highest level of cognition in the human brain. This is the part of the brain that allows for memory, perception, planning, reasoning, and dual tasking, or completing two tasks at the same time. Over the duration of many years of studying Parkinson disease, there have not been many studies done that compare the executive functioning of participants with Parkinson disease and those who are at a neurologically normal functioning level. The research study that I am conducting will allow for a comparison of the two groups so that those results can be taken and used as a basis for more research and better treatments. Thus far, not much research has been done on this particular topic so I will be looking to see if there is a difference in executive functioning in individuals with Parkinson and individuals who are neurologically normal.
The method that I am using to get my data is by using a Multiple Errand Test. The Multiple Errand Test is a simple test that will take place after the participant fills out a consent form and a demographic and medical history questionnaire. After that portion is completed, the test requires participants to gather a number of items from a grocery store in an allotted amount of time. This test will analyze the cognition, perception, memory, and dual tasking abilities in the participants.
The Multiple Errand Test allows examining all of these parts together and seeing what part Parkinson disease affects the most. This research is important because more than one million Americans live with Parkinson disease. This research will help produce better treatment plans for patients with Parkinson disease as well as gaining a better understanding of what this disease does to the mental capacity of individuals.
I will then take the results from the participants with Parkinson disease and the neurologically normal participants and compare them. This will help us come to a conclusion about the effects of Parkinson disease on executive functioning .
Based on prior research about the deterioration of cognition and the associated cognition processes due to Parkinson disease, I would expect to see a large gap in the executive functioning of those with Parkinson disease versus those who are neurologically normal. The detrimental effect of Parkinson disease disintegrates the ability to plan, to dual task, and to remember, leaving a long-term effect of dementia. Hopefully, this research study will help lead to new discoveries about Parkinson disease and better treatments for those who have it.
Rachel Fletcher, Microbiology & Molecular Genetics
Research: Inbreeding Depression and Cleistogamous Species
[View my work!]
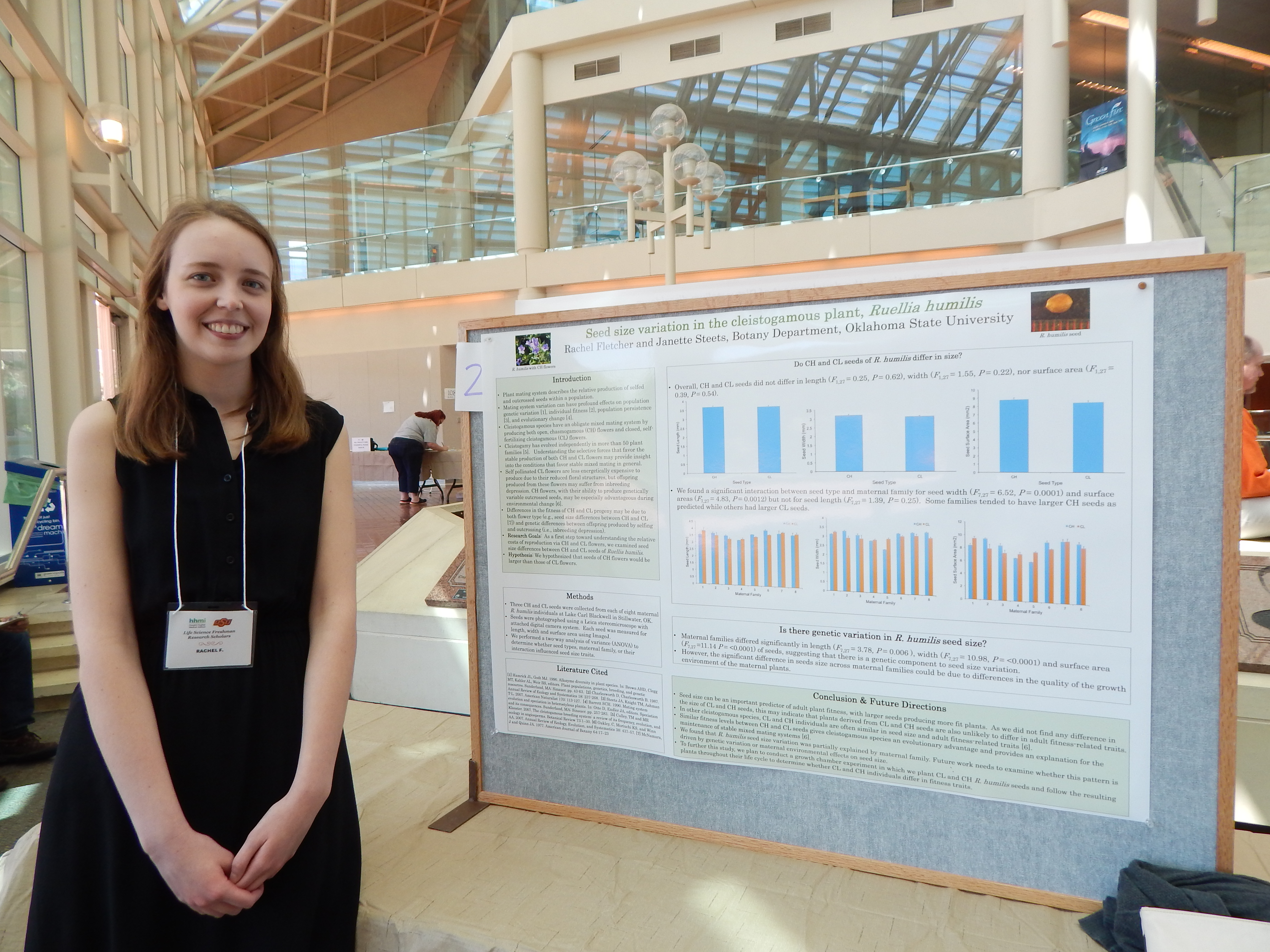
Project Summary: Most flowering plants are hermaphroditic, and thus, have the ability to self-fertilize or outcross. While some evolutionary theory predicts that only complete selfing or complete outcrossing are evolutionary stable strategies, a recent survey of flowering plants finds that over 40% of species have a mixed mating system (i.e., outcrossing rates between 20-80%; Goodwillie et al. 2005). Cleistogamous plant species have an obligate mixed mating system by producing heteromorphic flowers; a given plant produces a mixture of closed, selfing flowers (cleistogamous, CL) and open, showy flowers (chasmogamous, CH) that may be outcrossed or self-pollinated (Oakley et al., 2007). Cleistogamous species provide strong evidence for the evolutionary stability of mixed mating systems (Oakley and Winn, 2008).
There are fitness benefits and costs to the production of both flower types. Evidence suggest that CL flowers are energetically less expensive than CH flowers because they do not rely on a pollinator to produce fruit and are often smaller than the CH flowers (Oakley & Winn 2008). However, as the offspring produced from CL flowers are selfed, they may suffer reduced fitness due to inbreeding depression (Goodwillie et al. 2005). Thus, CL flowers are not able to adapt to change in the environment quickly since they have a limited gene pool. CH flowers, however, increase the genetic variation in a population by allowing for mating between individuals (Oakley & Winn 2008). Greater genetic variation among offspring produced by CH flowers may be advantageous during times of environmental changes, as genetic variation is the raw material for evolutionary change (Oakley & Winn, 2008).
In this experiment, we will use Ruellia humilis as a model system to explore whether seeds produced by CL and CH flowers differ in fitness-related traits.
We hypothesize that the seeds produced by CH flowers will be larger in size and have higher germination rates than those produced by CL flowers. In addition, we hypothesize that the resulting CH plants will be larger in size and have higher survival rates than CL individuals. We collected naturally pollinated CL and CH seeds from eight maternal R. humi/is individuals growing at Lake Carl Blackwell, Stillwater, OK. Seeds will be photographed using a Leica stereoscope and camera system. Seed size (length, width, surface area) will be quantified using image analysis software (Image J). To promote germination, seeds will be cold stratified for two months. Following stratification, they will be planted in pots filled with commercial potting soil and grown in a growth chamber for three months. Plants will be checked daily to note germination timing and survivorship. At the end of the growth period, plants will be measured for height, leaf size and leaf production. Statistical analyses will be performed to determine whether CL and CH offspring differ in any fitness-related traits.
Grace Flusche, Biochemistry & Molecular Biology
Research: Comparison of Two Adjuvants in an Experimental Actinobacillus Equuli vaccine in Horses
[View my work!]
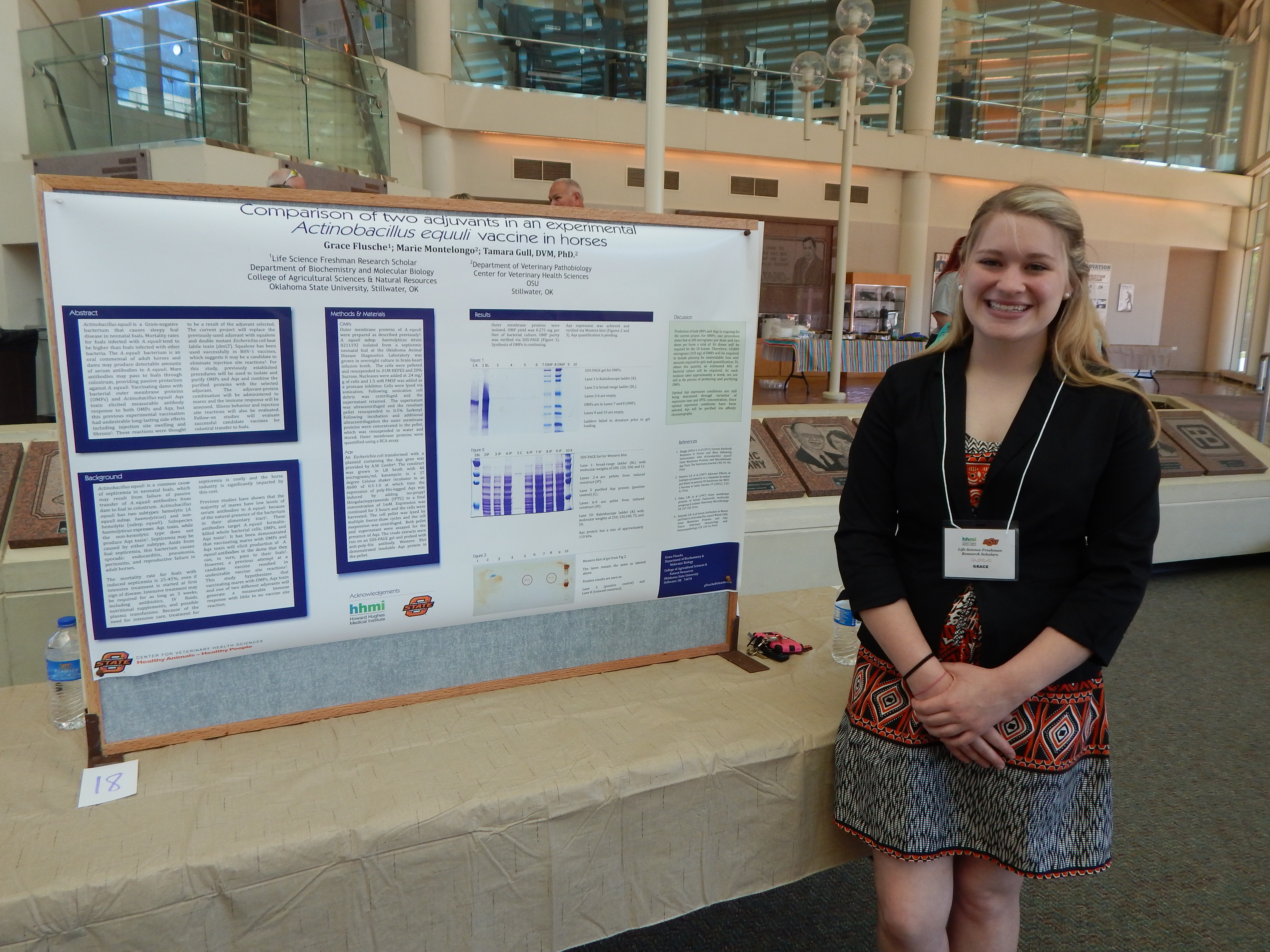
Project Summary: Actinobacillus equuli is a Gram-negative bacterium that causes sleepy foal disease, a septicemia found in neonatal foals. This disease starts with diarrhea, and can possibly lead to meningitis, pneumonia, purulent nephritis, or septic polyarthritis. Deaths are co=on in foals infected with A. equuli; mortality rates for foals with A. equuli are often higher than for other bacterial illnesses. This bacteria originates in the dam's mouth, respiratory tract, and alimentary system, and is passed to the foal during or innnediately following birth. Dams produce detectable amounts of antibodies to the outer membrane proteins (OMPs) of A. equuli and research shows they are able to pass this i=unity to their foals. These antibodies are naturally occurring in the dam's colostrum and have the ability to ward off the disease. It is hypothesized that this phenomenon can be aided by vaccinating the dam with OMPs and Aqx toxin, which belongs to the RTX family of pore-forming toxins that are toxic for horse leukocytes and erythrocytes. A vaccine has been created and tested for this disease, and although it boosted the innnunity of the foals, the adjuvant used was hypothesized to be the reason for injection site reactions- long lasting swelling and fibrosis that could not be prevented by any means of site preparation. Further research with different adjuvants in this vaccine is required to obtain a sustainable and effective vaccine that will be acceptable to horse owners.
This study will involve replacing the former adjuvants with squalene and double mutant Escherichia coli heat labile toxin (dmLT). Previous studies with squalene used in conjunction with sulfolipo-cyclodextrin for BHV-1 vaccines suggest these adjuvants to be a desirable solution to the injection site reaction elicited by other adjuvants; research involving using dmLT is still ongoing. To create the vaccine, we will begin by preparing cell envelopes via sonication.
Then we will extract and purify the outer membranes of A. equuli. We will manufacture Aqx toxin using an E. coli construct that contains a plasmid that can be induced to make the Aqx toxin. The toxin will then be extracted using histidine-tagged column extraction. We will repeat these processes until enough product is acquired. Each dose of the vaccine will be composed of 200 ug OMPs and 200 ug Aqx toxin combined with 50 ug of either squalene or dmLT, qs. to 1 mL with phosphate-buffered saline. The mares will be injected on Day 0 and again on Day 21 for a booster. We will collect serum samples from the mares on days 0, 7, 14, 21, 35, 42, 56, 70, 154 and 238 following the initial inoculation. The mares' injection sites will be observed daily for the first seven days, and weekly thereafter until Day 238. The serum samples will be assessed for antibodies against both A. equuli OMPs and Aqx toxin.
This modified vaccine is hypothesized to have similar immunity-boosting qualities for mares as found in previous studies, but less reaction site distress to their darns. The expectation for this study is to scale-down the side effects of reactions within acceptable limits, while maximizing the innnunity the foals receive, to create a vaccine that is safe and effective.
Jordan Gauss, Biochemistry
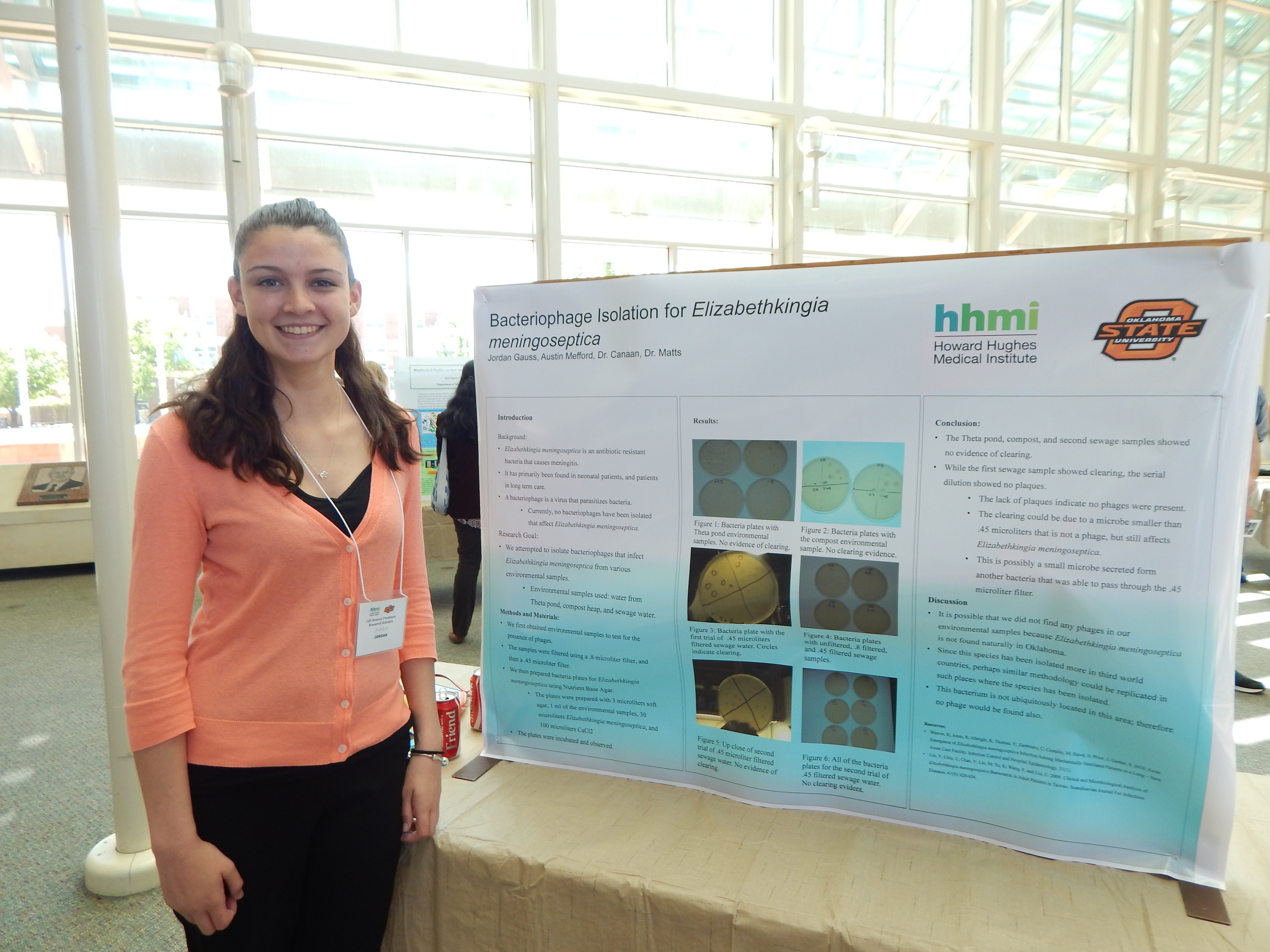
Research: Bacteriophage Isolation of Elizabethkingia meningoseptica
[View my work!]
Project Summary: Elizabehtkingia meningoseptica is an anti-biotic resistant strain of bacterial meningitis. Outbreaks of the strain of meningitis are common in hospitals, particularly in patients in long term care, and neonatal intensive care patients. Because this strain is resistant to most common antibiotics, infections with this strain of meningitis are almost always fatal. Aside from this bacteria's resistant to common antibiotics, very little is known about this bacteria. Inan effort to better understand this bacteria and prevent infections, we are interested in isolating bacteriophages that affect Elizabethkingia. We hypothesize that where we are able to isolate Elizabethkingia meningoseptica, we will also isolate a bacteriophage that affects Elizabethkingia.
To isolate the bacteriophages, we will first collect environmental samples that likely contain this strain of Elizabethkingia. Elizabethkingia thrives in many different environments. We will primarily use soil samples to isolate the bacteria. After obtaining the samples, we will liquefy the samples, so they will pass through our filtration system. We will make cultures of these samples to first isolate the bacteria itself, and determine whether the sample contains Elizabethkingia meningoseptica. The liquid samples will also be run through our filtration system that will isolate our bacteriophages. After any bacteriophages are isolated, we will then move to a different lab and test to see whether or not these bacteriophages infect Elizabethkingia meningoseptica. In addition, we hope to use microscopy to observe the form of these bacteriophages. However, we do not anticipate that the available microscopes will be of high enough power to properly observe the bacteriophage.
To ensure our filters isolate the bacteriophages, we have multiple filters of varying size. This is to ensure that our filters are not too big, so that the bacteria filters out. In addition, we have to ensure that our filters are not too large, so that we lose the bacteriophage in the filtration process. In addition, we are testing multiple protocol for the bacteriophage isolation, so we may ascertain the most effective means of bacteriophage isolation for these particular bacteriophages. We speculate that most of our lab time will be spent culturing bacteria and filtering our environmental samples. Isolating bacteriophages that infect Elizabetkingia will give future researchers more information about this particular strain of meningitis.
With our research, we expect hope to find environments in which Elizabethkingia thrives. In addition, we are hopeful that we will be able to isolate bacteriophages to which Elizabethkingia is susceptible. This would provide researchers with more information about Elizabethkingia meningoseptica, which could eventually lead to new ways of preventing outbreaks of this strain in hospitals. While we would like to be able to view our bacteriophages, as previously mentioned, this may not be possible with our resources.
Tabitha Gunnars, Kalyn Dealy, Courtney Manly, Zoology

Research: Shrews in a bottle
[View our work!]
Project Summary: We will investigate the relationship between two groups of small mammals (shrews, Soricidae , and mice and rats, Cricetidae) and roadside litter along with other variables. Small mammals have been found in bottles and cans in places such as England and Indiana, but use of roadside litter by small mammals has not been investigated in plains habitats, which are found in Oklahoma. The question we are investigating is why small mammals nest in roadside litter and what might cause them to do so. The variables we are observing are the vegetation patterns, weather conditions, and physical aspects of the types of litter in which small mammals are found.
We will sample a one mile long segment of Highway 177 between 32nd and 44th streets every two weeks between the last week of February and the first week of April. We will collect roadside litter along the road and examine it for the presence of small mammal remains and nesting material. Tabby Gunnars will be investigating the relationship between the occurrence of small mammals in litter and vegetation by sampling 20 randomly selected 1-m' plots along the highway and at every point where a small mammal is found in roadside litter. She will record the number of stems and whether plants are grasses, forbs, or woody vegetation.The reason vegetation will be observed is because of the effect it could have on the occurrence of small mammals. For example if there is plenty of vegetation cover, then there may be fewer small mammals using roadside litter for shelter. Kalyn Dealy will be monitoring weather data for her part of the study. She will use weather variables available from Mesonet, a website that receives local weather data from a station. She will be using the station two miles west of Perkins, Oklahoma. She will collect her data the day of and five days prior to sampling. Weather data may include whether it is cloudy or sunny, rainy, snowy, or the temperature. Analyzing weather data can help to determine if it affects when small mammals will go into roadside litter for shelter. Courtney Manly will be measnring the sizes, shapes, and colors of bottles and cans and the material (plastic, glass, metal, and paper) and comparing which types of containers were used by small mammals compared to containers that were not used. This type of data will help determine if small mammals are more attracted to some types of litter compared to others.
We expect to find more small mammals using roadside litter early in our study while the weather is colder and there is less vegetation to be used for shelter. We also expect to find more remains (skeletons) in bottles with smaller openings because it will be more likely for animals to get trapped.
Daniel Henthorn, Biochemistry & Molecular Biology
Mentor: Dr. John Gustafson, Biochemistry & Molecular Biology
[View my work!]
Project Summary: Staphylococcus aureus is a human opportunistic pathogen that harbors numerous virulence factors and can readily acquire genes that allow the organism to impede the effects of antibiotics (11, 8). One of the better-known examples of antibiotic-resistance mechanisms in S. aureus is methicillin-resistance (8) and in 2011, methicillin-resistant S. aureus (MRSA) led to 80,000 infections and 11,000 deaths within the United States (1).
Fusidic acid is a steroid antibiotic that was clinically introduced in the 1960s to treat S. aureus infections and is currently used widely throughout Europe and the commonwealth and is soon to be licensed for use in the United States (9, 5). Fusidic acid mode of action is to act as a protein synthesis inhibitor (2, 3).
However, fusidic acid resistance is a rising issue in many places. This resistance is characterized by two distinct ways: The first is through a naturally-occurring mutation in the gene fusA. (7) The second is by the acquisition of fusB and fusB homologues (10, 6).
Our laboratory previously discovered two genes that were induced in the presence of fusidic acid (4) and are here named as fusidic acid induced A and B (faiA and faiB). Both of these genes are thought to code for a bipartite multidrug efflux pump. Upstream of faiA lies the divergently encoded tetR homologue, faiR, and we hypothesize that FaiR controls the transcription of faiAB.
In order to have adaptable survival in an unstable environment bacterium have adaptive responses triggered by regulatory proteins that, in response to environmental signals, altar transcription (12). One large family of these regulators is the TetR family, originally identified as a regulator of the tet genes, which confer resistance to tetracycline (12). In S. aureus FaiR in understood to be a TetR homologue regulating faiAB, which codes for a putative fusidic acid drug efflux pump.
In this project we will determine if the deletion of faiR will affect the level of fusidic acid resistance expressed by a fusidic acid-selected fusA mutants of strain JE2faiR constructed in our laboratory. We hypothesize that since faiR acts as a negative regulator of faiAB expression, JE2faiR-fusA mutants will express a higher level of fusidic acid resistance as compared to JE2fusA mutants.
Methods:
JE2 is a community-associated MRSA strain, cured of all plasmids, which will be utilized for this study. We will first select for JE2 and JE2ΔfaiR fusA-mediated fusidic acid-resistant strains. To do so we will start by taking an overnight culture of both strains and plating them onto Muller Hinton agar (MHA) plates that contain 2 μg/ml fusidic acid and incubate these plates overnight at 37°C. We will pick 6 colonies and streak them onto drug free MHA and incubate them overnight at 37°C. This will be done twice. We will then use minimal inhibitory concentration (MIC) and antimicrobial gradient assays to determine the levels of resistance/susceptibility to fusidic acid.
We hypothesize that since faiR acts as a negative regulator of faiAB expression, JE2faiR-fusA mutants will express a higher level of fusidic acid resistance as compared to JE2-fusA mutants. This data will be seen in both in both the MIC and the gradient plates. However, another possibility is if faiR is a positive regulator of faiAB expression, then we would expect JE2faiR-fusA to have a lower expression of fusidic acid resistance. We do not know which is true of faiR, and the results of this experiment would help expand what we do know of this regulator gene.
Jaelen Herron, Biology

Research: Study of Annual Rings in Native Herbs in Stillwater, Oklahoma
[View my work!]
Project Summary: In the experiment, we are conducting research at McPherson preserve. This preserve is in Stillwater, Oklahoma and is owned by Oklahoma State University. This system is used to conduct scientific research on natural conditions and is on over 2000 acres. The experiment is answering the question what herbs, specifically perennial herbs, in Stillwater, Oklahoma have perennial rings. The experiment will address the number of perennial herbs in Stillwater that demonstrate annual growth rings. There has not been much research conducted on these perennial herbs and annual growth rings.
The McPherson preserve is home to many herbaceous species. These herbaceous species will be excavated and taken to the laboratory. At least one of each herbaceous species will be excavated for research. In the laboratory using a microtome the herbaceous species will be sliced into 15-30 µm thick slices. After the herbaceous species are sliced they will be thickened with absolute alcohol to harden the tissue of the roots that may be relatively soft. The cutting will
then be permanently dyed with safranin. The cutting will then be put onto a microscopic slide and examined for annual growth rings.
Similar experiments have been conducted on perennial herbs and annual rings in locations such as Michigan, Europe, etc. An experiment in Michigan conducted by Hansjorg Dietz and Fritz Hans Schweingruber with similar motives has produced successful results on the correlation of some perennial herbs and annual growth rings. The experiment is similar in theory, but in different locations with very different climate conditions. The approach of going into locations to collect perennial herbs and analyze annual growth rings has proven to be highly effective.
The experiment is expected to discover 12-15 species to be identified in winter conditions. Over all we expect to find a plethora of perennial species that have annual growth rings. Each ring will vary in thickness based upon the amount of rain during the year. The annual growth rings will also depict the age of each perennial herbs. The results from the experiment will to establish a broader basis for the applicability and the limits of herb chronology.
In a side experiment investigations are being conducted where starch is predominately located in the secondary xylem of the root collar during the winter months. This matters because we are investigating whether if all the starch is located in only the most recent annual rings or if it is allocated throughout all rings while also looking to see in what cell types the starch is located. It is suspected that how much starch is stored during the course of the growing season will also affect the width of the year's annual ring.
Kimberly Keck, Zoology
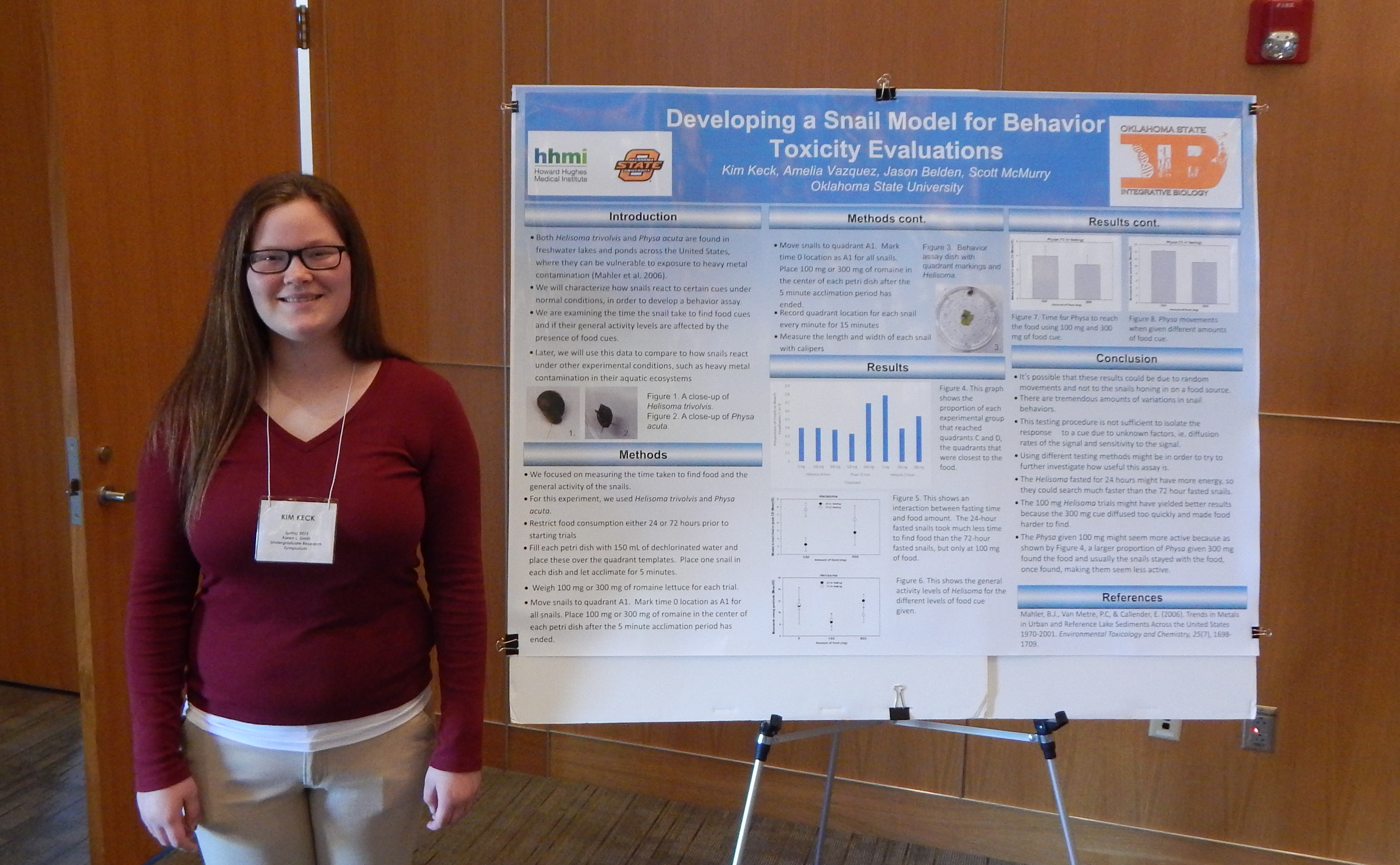
Research: Lead Accumulation and Toxicity to Snails
[View my work!]
Project Summary: In Picher, 0klahoma, sits the Tar Creek Superfimd site the most toxic site in the United States. Tar Creek owes its existence to a lead mining boom from 1913 to 1967 in which the Picher mines were the nations largest producers of lead and zinc, indeed one of the largest in the world. When mining ceased, mines were abandoned and chat piles dominated the landscape. Eventually, the mine shafts filled with water that became contaminated with heavy metals and leaked out into surrounding water sources (e.g., Tar Creek). The chat piles were also a source of heavy metals into water sources and terrestrial environments via contaminated dust.
Scientists can measure the amount or accmru.ilation of contaminants, like heavy metals, in organisms to assess the bioavailability as part of the overall assessment of exposure and effects at contaminated sites. Choosing the correct organism to study is important, and is based on certain criteria. For example, diet, movement patterns, abundance, and other fuctors are important considerations. Snails are often an abundant organism in aquatic systems and do most of their fucding in relatively small areas. These characteristics make snails a potentially useful sentinel species in field studies. Unfortunately, not enough is known about how heavy metals (lead in this case) accmnulate in snails and their shells to use as a marker of contamination and bioavailability in a system, like the Tar Creek Superfund site and its assorted drainages. Inthis study, I will examine how the accmnulation oflead in snails and their shells and the corresponding effects on toxicity to the snails.
I will use the juvenile bladder snail (Physa sp.) from an existing culture in Jason Belden's Jab at Oklahoma State University. Snails will be kept in gravel-lined tanks filled with aerated water and fed the same amount of broccoli each day. The snails will be maintained in aerated tanks with gravel along the bottom
The experimental design will consist of four lead concentrations replicated three times. Each tank replicate will contain ten snails, with the tanks set up in a three-by-four pattern on one shelf with one replicate per row and the positions of each tank in the row randomly selected. Treatments will consist of lead-free water (contro, low lead, medirnn lead, and high lead. The medinm lead will be determined from previous literature that set a lowest observable effective concentration oflead for snails to be in, with the low concentration set at half of the medinm concentration, and the high concentration set at twice the medirnn concentration.
Snails will be exposed over 21 days and mortality measured every 24 hours. Snail mass willbe determined at day zero fur all snails and day 21 for the surviving snails (±1 rng). Dead snails and snails remaining after 21 days will be removed from the tanks, labeled with lead concentration, replicate number, the date of death, and individually stored at -20°C. They will then have their soft tissue removed and shells digested in acid prior to analysis for trace metals.
Previous research indicates that some snail species exhibit low death rate in elevated metals levels, presumably due to their ability to store metals in their shells, rendering it U11available for toxic effects. This study will show the accrnnulation of lead and mortality over time. It will give us needed information about the effects of exposure time and lead concentrations on the toxicity to snails.
Sam Kimbrough, Microbiology & Molecular Genetics
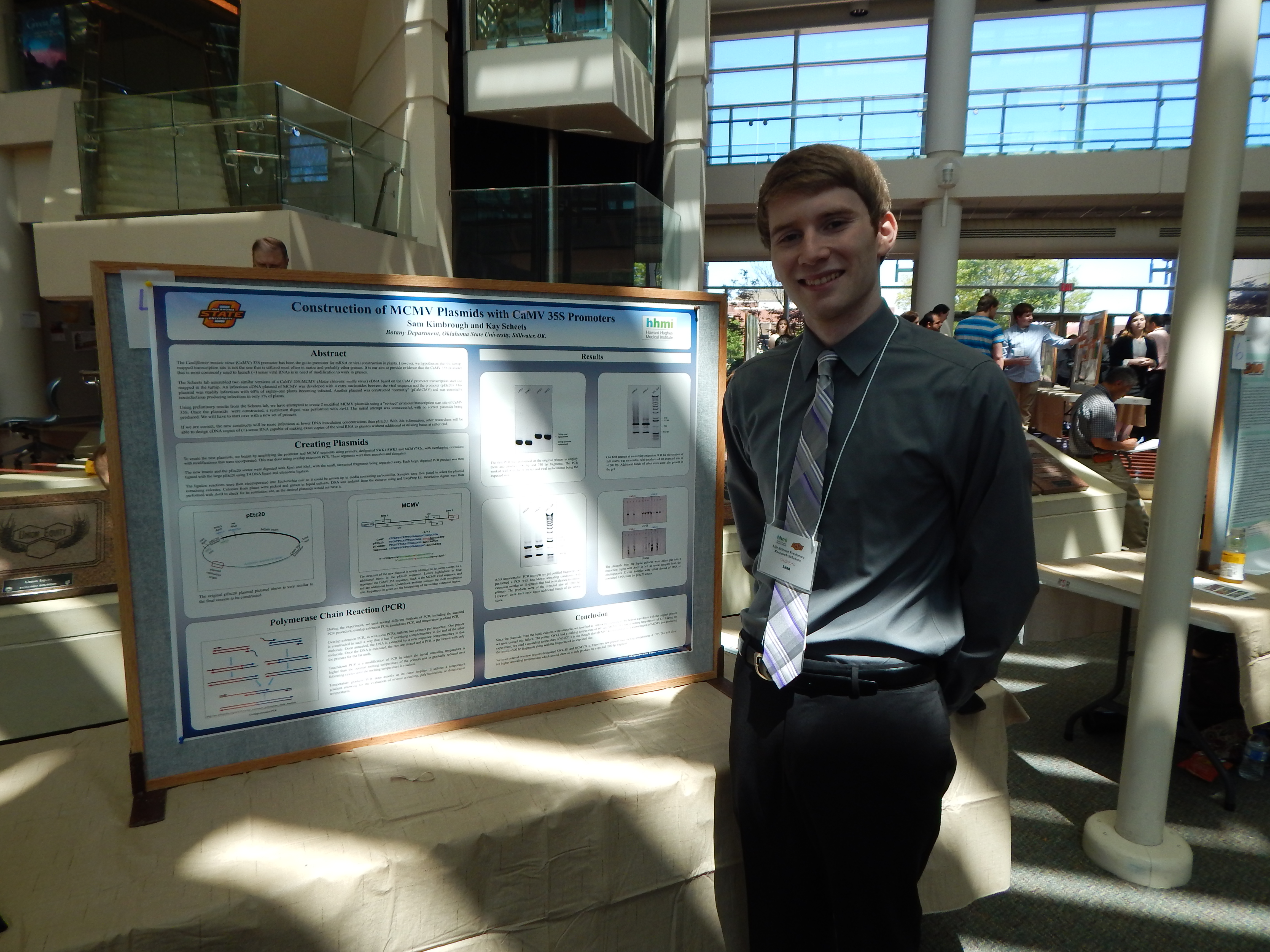
Research: Construction of MCMV Plasmids with CaMV and ScBV 35S Promoters
[View my work!]
Project Summary: The Cauliflower mosaic virus (CaMV) 35S promoter has been the go-to promoter for mRNA or viral construction in plants. However, we hypothesize that the turnip-mapped transcription site is not the one that is utilized most often in maize and probably other grasses. It is our aim to provide evidence that the CaMV 35S promoter that is most commonly used to launch (+) sense viral RNAs is in need of modification to work in grasses.
The Scheets lab assembled two similar versions of a CaMV 35S:MCMV (Maize chlorotic mottle virus) cDNA based on the CaMV promoter transcription start site mapped in the turnip. . An infectious cDNA plasmid of MCMV was developed with 4 extra nucleotides between the viral sequence and the promoter (pEtc20). This plasmid was readily infectious with 60% of eighty-one plants becoming infected. Another plasmid was constructed “normally” (pCaMCMV) and was essentially noninfectious producing infections in only 1% of plants.
Using information from the Scheets’ lab preliminary results, new MCMV with a “corrected” promoter/transcription start site junction of CaMV 35S will be made. Maize seedlings will then be rub-inoculated. The test will involve positive controls, negative controls that will be mock inoculated, and the test plasmid group. The plants will be examined daily for symptoms. Small leaf samples will be collected at three weeks and assayed for MCMV. The assay will be done with rabbit antisera raised against MCMV virions using an ELISA assay.
If we are correct, the new constructs will be more infectious at lower DNA inoculation concentrations than pEtc20. With this information, other researchers will be able to design cDNA copies of (+)-sense RNA capable of making exact copies of the viral RNA in grasses without additional or missing bases at either end.
Jamie Landers, Zoology

Research: Determining Sexual Maturity of Crotaphytus collari hatchlings and yearlings as compared to body size, sex, and season
[View my work!]
Project Summary: Sexual maturation varies across all species. In humans, it generally happens between the ages of 10 and 15 years of age. In most other animal species, sexual maturation occurs when the animal reaches adulthood. In a few animal species, sexual mature is linked to body size. These species will be sexual mature once they hit a certain size regardless of their age. This type of sexual maturity is what I am setting out to test in Crotaphytus collaris, eastern collared lizards. Collared lizards are special in that they display sexual dimorphism. Even as hatchlings, males have bright, horizontal orange bars that reach across their backs while females’ coloring remains drab.
The purpose of the hatchling orange bars is still up for debate but the questions the orange bars raise are vital in understanding their purpose. Collared lizards also practice the act of pair bonding. Pair bonding is where males of the species spend time with females to create a bond so that when mating season comes, the females will be more likely to join a particular male’s mating harem. A harem is a group of females who mate with a particular male collared lizard and stay in that male’s territory. Collared lizard hatchlings also practice pair bonding and territorial instincts although they are not thought to be sexually mature. This raises the question of why would the hatchlings exhibit these behaviors if they are not yet sexually mature. To test this question, I will be collecting a cloaca smear from yearlings in the spring and hatchlings in the spring. This involves expressing the cloaca of the specimen onto a microscope slide and letting the slide air dry.
Then back in the lab, I will look at each slide under a microscope to observe if the slide has the presence of mature spermatozoa. If mature spermatozoa are present, the specimen lizard is sexually mature. In order to obtain the cloaca smears, I will travel with Dr. Fox and his graduate students to the dam site, SL1, at sooner lake. The field steps of the project are as follows: capture specimen, record the area the specimen was found, identify specimen, take body measurements, and perform a cloaca smear.
After capturing the specimen, I will record the specific area of the SL1 site where the specimen was found. This step helps in identifying trends in results and to keep data organized. Each lizard, apart from newborn hatchlings, has a set of toe clippings and sometimes paint drops on the back to identify it. Identifying the specimen used in my study will prevent errors in age and sex when recording and comparing data. Body size measurements are taken to compare to sexual maturity. The measurement is taken from nose to vent on the collared lizard. The cloaca smears will be compared to body size, sex, and season to produce the results of the project. We are expecting and hoping that the hatchings will not turn out to be sexually mature so that we can continue investigating the reason behind the hatchling orange bars.
Cerra Linn, Biology

Research: Purification of the INI1 Protein
[View my work!]
Project Summary: Atypical Teratoid/Rhabdoid tumor (AT/RT) is a tumor found in the central nervous system in infants usually under the age of 3. It is thought that development of AT/RT is correlated to the loss of the protein SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 (SMARCB 1), a tumor suppressor gene more commonly referred to as INI1 (Childhood Central Nervous System, 2014). lNI1 is a part of the SWI/SNF protein complex that regulates transcriptional activity through chromatin remodeling. Changes in genes such as INI1 may lead to cancer due to the chromatin remodeling taking place (Birks, 2008). ). These mutations are what cause the rapid growth of the AT/RT tumors and can either be spontaneous or be inherited from the infant's parents. They generally have no known cause inwhat makes them appear and cannot be prevented if the child has inherited it or if the gene has mutated. Previously AT/RT was thought to be a type of Medulloblastomas - another type of brain cancer - (Medulloblastomas, 2012) but is now treated as a separate tumor with different treatment options (AT/RT - Atypical Teratoid Rhabdoid Tumor, 2014).
The basis of this project will be to purify the INI1 protein and gain a better understanding of its structure and see how a purified sample of INI1 can be obtained by using methods of purification. This fist includes obtaining our sample of DNA from E.coli bacteria cells that will be expressed with the INI1 protein. Then, we will use polyhistidine-tag purification method which will allow for the lNI1 proteins to bond to the Ni-NTA allowing for the protein to be extracted and looked at alone. After this a SDS-PAGE gel will be run in order to assess the purity of the protein. Ifthat protein is pure then the final elute product should have only one band at about 47, 000. If a purified sample is obtained, the sample will be used to try to get a crystal of the protein by using x-ray crystallography. lf this is achieved we will be able to see how the purified structure is similar to other proteins and may be able to learn more about how it forms the AT/RT tumors and what may trigger it to develop.
Through this purification process we hope to successfully isolate the INI1 protein. However, this process will require many trails and errors because we do not know the best method on how to purify this protein and if it will even work. We expect to successfully purify the protein this semester and then move on to x-ray crystallography this corning fall. We hope that by accomplishing this it will allow us to know how the INI1 protein plays a part in the development of the AT/RT tumors and how it may be prevented or reversed.
Rachel McAllister, Microbiology & Molecular Genetics
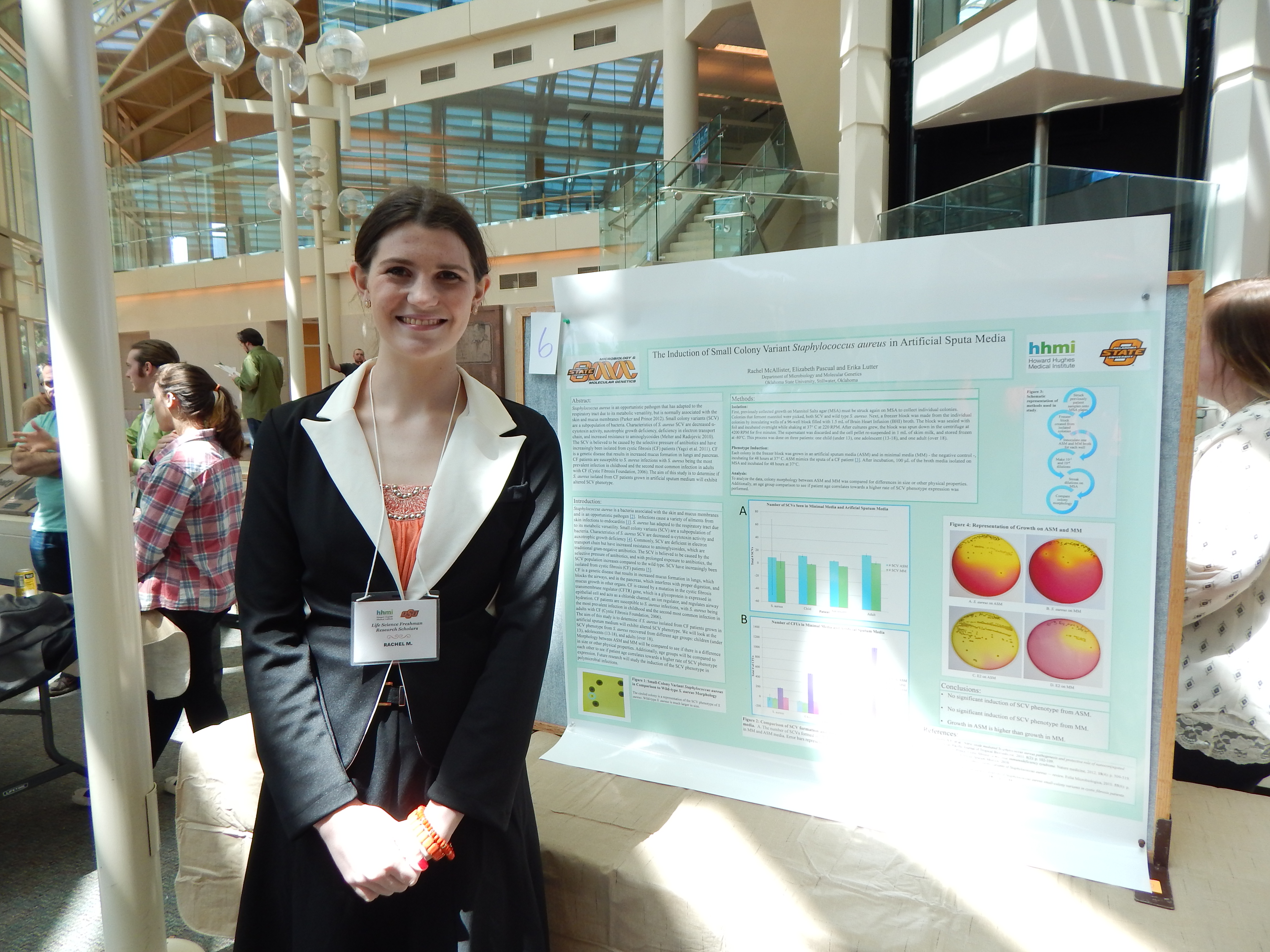
[View my work!]
Project Summary: Staphylococcus aureus is a bacteria associated with the skin and mucus membranes and is an opportunistic pathogen (Parker and Prince 2012). Infections cause a variety of ailments from skin infections to endocarditis (Chakraborty 2011). S. aureus has adapted to the respiratory tract due to its metabolic versatility. Small colony variants (SCV) are a subpopulation of bacteria. Characteristics of S. aureus SCV are decreased a-cytotoxin activity and auxotrophic growth deficiency (Melter and Radojevic 2010). Commonly, SCV are deficient in electron transport chain but have increased resistance to aminoglycosides, which are traditional gram-negative antibiotics. The SCV is believed to be caused by the selective pressure of antibiotics, and with prolonged exposure to antibiotics, the SCV population increases compared to the wild type. SCV have increasingly been isolated from cystic fibrosis (CF) patients (Yagci et al. 2011).
CF is a genetic disease that results in increased mucus formation in lungs, which blocks the airways, and in the pancreas, which interferes with proper digestion, and mucus growth in other organs. CF is caused by a mutation in the cystic fibrosis transmembrane regulator (CFTR) gene, which is a glycoprotein expressed in epithelial cell and acts as a chloride channel, an ion regulator, and regulates airway hydration. CF patients are susceptible to S. aureus infections with S. aureus being the most prevalent infection in childhood and the second most common infection in adults with CF (Cystic Fibrosis Foundation, 2006).
The aim of this study is to determine if S. aureus isolated from CF patients grown in artificial sputum medium will exhibit altered SCV phenotype. We will look at the SCV phenotype from S. aureus recovered from different age groups: children (under 13), adolescents (13-18), and adults (over 18). First, previously collected growth on Mannitol Salts Agar (MSA) must be struck again on MSA to collect individual colonies. Both SCV and wild type S. aureus will be selected.
Next, a freezer block will be made from the individual colonies by inoculating wells of a 96-well block filled with 1.0 mL of Brain Heart Infusion (BHI) broth and incubating overnight while shaking at 37° Celsius at 220 RPM. After cultures have grown, the block will be spun down in the centrifuge at 4200 RPM for five minutes. The supernatant will be discarded and the cell pellet re-suspended in 1mL of skim milk, and stored frozen at -80°C. The negative control is growth and morphology on a minimal media (MM). One colony will be grown in an artificial sputum media (ASM) and in MM, incubating for 48 hours at 37° Celsius. ASM mimics the sputa of a CF patient. After incubation, 100 µL of the broth media will be plated on MSA, and incubated for 48 hours at 37°C.
To analyze the data, morphology between ASM and MM will be compared to see if there is a difference in size or other physical properties. Additionally, age groups will be compared to each other to see if patient age correlates towards a higher rate of SCV phenotype expression.
Brienna Milleson, Microbiology & Molecular Genetics

Research: Octoploidy-induced Genome Downsizing and its Effects on Plant Morphology in Arabidopsis thaliana
[View my work!]
Project Summary: Arabidopsis thaliana, a model organism for plant genetics, is known to be capable of experiencing increases in ploidy, or the number of chromosome sets. This is due to the formation of unreduced gametes as a result of errors in chromosome pairing or unbalanced chromosome segregation in meiosis I. The TARDY ASYNCHRONOUS MEIOSIS (TAM) gene is responsible for positively regulating cell cycle progression in Arabidopsis meiosis. The TAM mutants display delayed and asynchronous cell divisions during male meiosis, producing unreduced gametes by skipping meiosis II entirely. This process of polyploidization continues until it reaches octoploidy and becomes unstable. The progeny of octoploid tam-2 are of reduced ploidy as a result. This purpose of this study is to examine fertility and morphological changes over at least two generations from the octoploid tam-2 variant of A. thaliana.
We expect that the instability of the genome may result in new genome compositions that produce new phenotypes in these generations. To investigate this, we will grow tam-2 octoploid mutants, along with wild-type diploid, tetraploid, hexaploid, and octaploid varieties of A. thaliana for comparison of morphology according to ploidy level. During growth, cotyledon samples fixed in ethanol and will be taken for examination using light microscopy. The samples will be stained with 4'.6-diamidino-2- phenylindole (DAPI) to determine the nuclear size in guard cells using fluorescence microscopy. The series of guard cell nuclear sizes corresponding to the different ploidy levels will serve as the standard values for determining the ploidy levels in the progenies of the octoploid tam-2, as well as for the creation of a growth curve from the data. Fertility for each generation will be investigated by counting the number of seeds per silique.
We will use a compound microscope and a dissecting scope equipped with a digital imaging system to detect morphological changes at the cellular level. If we observe significant phenotypic changes, we will use the polymerase chain reaction technique, or PCR, to investigate genomic changes and attempt to identify the mutations responsible for the phenotypic changes. We will use 0.8% agarose gel matrix stained with Phenix's GelRed Nucleic Acid Stain for electrophoresis to digitally detect the length of the PCR products. Through this, we may observe how often these changes occur in a given ploidy level and generation. If deemed necessary, we will also employ one-step Reverse Transcriptase PCR, or RT-PCR, techniques to detect changes in the transcription of certain genes. This study will be the first time that the process of morphological change due to genomic decay can be observed in real time in the lab.
The findings of this study should have important implications to the mechanisms for angiosperm speciation, and could be extended to aid in designing techniques to engineer new crops in the agricultural industry.
Joshua Moore, Microbiology & Molecular Genetics

Research: G-Alpha 8 Phenotype Identification in Differentiation
[View my work!]
Project Summary: Dictyosteliurn Discoideum is both a singular and multi-celled organism and is primarily found in soil samples around the world. Dictyostelium is an amoeba widely used in scientific molecular biology and genetics research because of its ability to communicate between its sovereign cells within the organism to move systematically as a single unit while still maintaining the ability to divide into distinct entities as well as to fuse two self-reliant entities together into a single system when necessary. Scientists use this organism to study everything from signal transduction between the different cells to the genetics of the entire population. This allows Dictyosteliurn Discoideum to be incredibly flexible and useful in many different types of research.
Specifically I am analyzing the G- alpha 8 protein located in Dictyosteliurn cells. G- alpha 8 is a specific type of GTP proteins which are vital to the functionality of signal transduction pathways which are utilized in aggregation of the cells to form a single aggregate when the cells become starved. After the cells join together they then go through a fruiting body process where they then the majority of the cells transform into a vegetative state to conserve their limited energy and resources and lie dormant until resources become favorable again. The GTP proteius are vital to the communication between cells in the aggregate and the formation of the final fruiting bodies to enter a dormant state. Intotal there are known to be 12 different G-alpha proteins; the majority of these proteins phenotypes and functions have been identified and defined in the aggregation process done through other studies except for G- alpha 8, and one or two others.
Currently, G-alpha S's phenotype and role in the aggregation process have not been identified aside from several unsupported or proven speculations previously done by Dr. Wu and his colleagues from John Hopkins University. So, what is the G-alpha 8 role in the aggregation process of Dictyostelium? Thats is the question I am hoping I will be able to answer; currently the answer to the question is still very unclear. To approach this question Dr. Hadwiger has given me a process he which he thinks may lead to answers. In my experiment I have obtained a wild and natural strain of Dictyostelium cells and prepared a stock culture of cells in our standard HLS media solution, which allows the cells to more than enough resources to avoid the aggregation step. To analyze G-alpha 8 I have prepared a small scale plasma prep specifically with antibiotic resistance and green fluorescence; the resistance will allow us to be able to grow a mutated culture of Dictyostelium with fluorescence present in the G-alpha 8 protein.
To create the mutant necessary to view this protein we used electroporation to increase the permeability of the cells and allow the plasmid to entire the cells. Unfortunately not all of the normal cells will be exposed to the plasmid; so I am using a G-1418 antibiotic to kill of all of the cells that do not possess any of the new DNA while still allowing the mutated cells to grow and continue to reproduce. After enough of the mutant cells have been grown we will then plate them on a non-nutritious potassium plate to force the Dictyostelium cells to entire the aggregation process. Then once the aggregation has begun we will monitor the movement of the fluorescent G-alpha 8 protein under a blue UV light under a microscope and observe and take pictures of the movement of the protein throughout the aggregate.
While we are not sure of the actual function and phenotype of Dictyostelium we expect that this experimental process will show that the G-alpha 8 proteins move to a particular part of the aggregate during the aggregation process; and from this we believe we will have enough information to form a possible answer to the function of the G-alpha 8 protein in the aggregation process of Dictyostelium Discoideum cells.
Maggie Pearce, Physiology
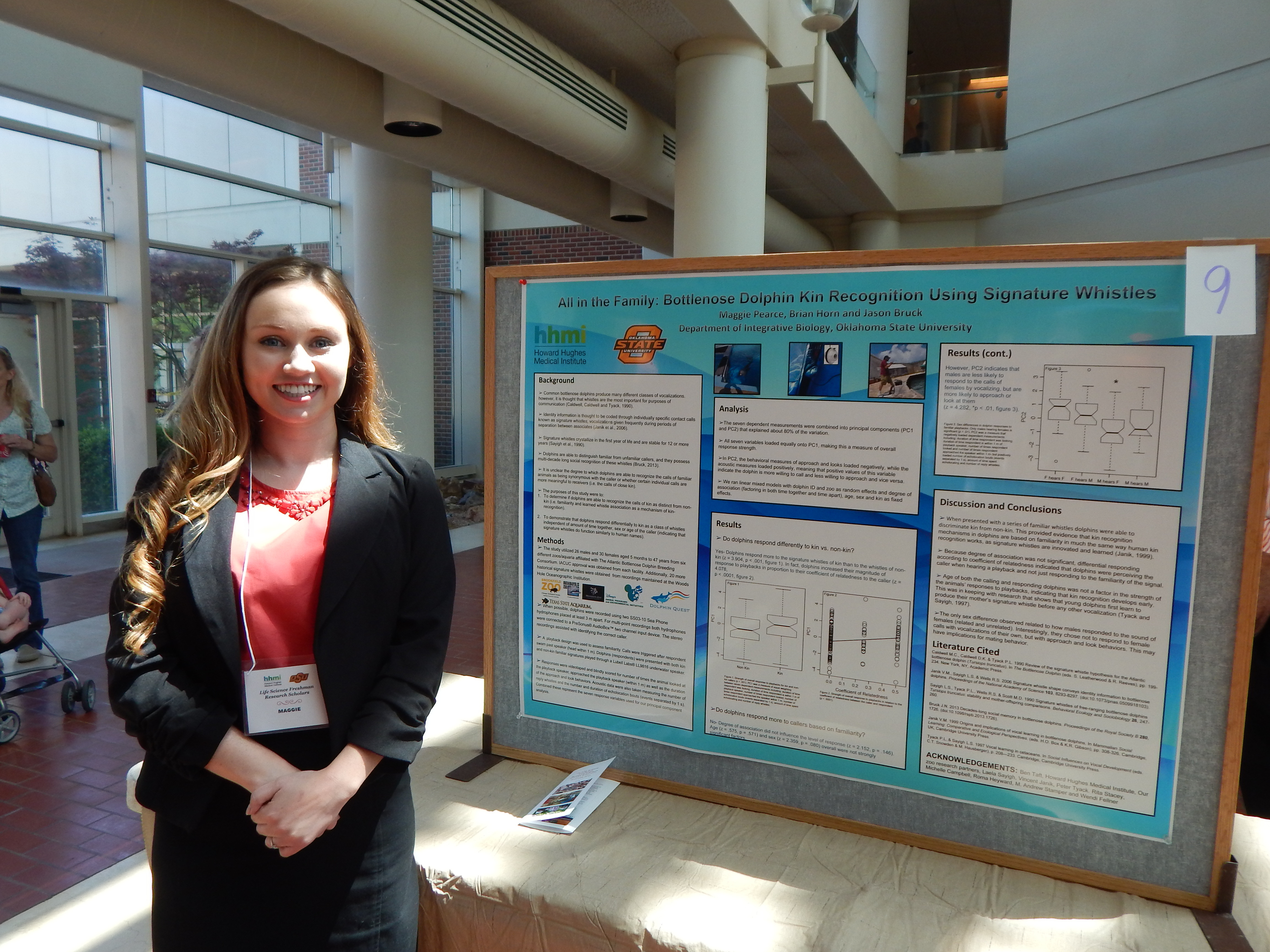
Research: Acoustic Kin Recognition in Bottlenose Dolphins
[View my work!]
Project Summary: Kin recognition is beneficial in preventing incest as well as determining which conspecifics to compete with and which to give aid to. In addition, the ability lo identify kinship encourages social growth and complexity. In the animal kingdom, there are various forms of kin recognition, such as phenotype matching in which an animal collects in formation on their own traits as well as those of familiar kin and uses the memory as a comparison. Bruck (2013), a previous study of recognition in bottlenose dolphins using signature whistles, showed that dolphins could recognize familiar dolphin whistles from unfamiliar dolphin whistles, but the ability to differentiate between kin and familiar nonkin has yet to be investigated. In order for a bottlenose dolphin to differentiate between kin and familiar non-kin, they must possess a familiarity based form of kin recognition, which is a cognitively more demanding form of recognition as compared to phenotype matching because it involves such processes as learning, memory and individual recognition because dolphin signature whistles are not determined by genetics but are developed by each dolphin individually in the beginning stages of life.
Our question for this study is, "Can dolphins differentiate between the signature whistles of kin and farniliar non-kin?" This study will test acoustic kin recognition in Bottlenose dolphins (Tursiops truncates). We will test this by using the recordings of dolphin whistles from Bruck (2013), and we will measure the response of dolphins to the signature whistles of kin and to those of familiar non-kin. The dolphins in the initial study were a part of a six institution breeding program, and their ages ranged from four months to forty-seven years. When we evaluate the videos of the 56 dolphins, I expect that the dolphins will show an increased response toward the playback of kin whistles than to those of familiar non-kin as a result of familiarity.
Lindsey Purcell, Aaron Seal , Biochemistry & Molecular Biology

Research: Cloning and Characterization of E.meningoseptica Beta-lactamases
[View our work!]
Project Summary: Throughout the semester, many studies will be performed on a bacterium named Elizabethkingia meningoseptica. It is considered to be a resistant to multiple antibiotics and produces a tissue called a biofilm, which is when bacteria coagulates to itself and forms a film similar to the plaque on the surface of your teeth by a method known as extracellular polymeric synthesis. The study bacterium is known to cause infections such as pneumonia, endocarditis, postoperative bacteremia, and meningitis in newborns and susceptible adults. The mortality rate when infected is more than fifty percent. Little is known about its antibiotic resistance or which antibiotics are more effective than others. It shows a great resistance to most antimicrobials that are used to treat gram-negative infections and is considered to be the reason for our research.
Given that there is little knowledge on the antibiotic resistant genes in Elizabethkingia meningoseptica, there is sufficient genomic sequence data that shows evidence that specific genes are responsible for the antibiotic resistance, however the correlation between genotype and phenotype has not been made. Targeting the predicted resistance genes is important and is considered a first step in the process. We have targeted these genes already, and we are designing primers that help this gene replicate in order for us to further characterize the gene as beta-lactamase. Beta-lactamase genes are what is considered to be the primary antibiotic resistant genes in Elizabethkingia meningoseptica confirming resistance to penicillins. Once amplified, cloned, and separated from other genomic data, we will clone the PCR product into a more known E. coli strain and test its insertion by DNA mini-prep for confirmation. We will then screen the clone strains for particular antibiotic resistance using the MIC/Kirby Bauer disk diffusion technique.
The goal in the research is to demonstrate a variety of antimicrobial resistance that E. meningoseptica exhibits. Using these tests will help in predicting various levels of susceptibility that the bacterium resists antimicrobials looking at the job of Beta-lactamase proteins. Using the MICs and MBCs we can also further isolate the hypersensitive and/or hyper-resistant strains of E. meningoseptica. Cloning antibiotic resistant genes found in Elizabethkingia meningoseptica into E. coli for comparison and characterization is vital to determine if any of these genes are susceptible to any antimicrobials. When antimicrobials are discovered, they may be a good option for commercial use. That of course will require more testing. This experiment will hopefully provide sufficient evidence to take the project in a forward direction including more research on the RND-type efflux pumps also found in Elizabethkingia meningoseptica that help provide more resistance to antibiotics. Where the development and characterization of the genes lead to a putative solution.
Jake Richmond, Zoology

Research: Determining Female Preference of the Collared Lizard, Crotaphytus collaris: Through Sexual Selection of Drab and Vivid Males
[View my work!]
Project Summary: All species have a varying preference for phenotypic traits for sexual selection. Sexual selection can be based on the specie's sexual dimorphism such as their color, size, and structure. A lot of the extraordinary morphological and behavioral diversity in the animal kingdom has come up through sexual selection. The species in particular that we are researching over is the Crotaphytus collaris along with their choices for sexual selection. Males of this species show off vivid orange bars horizontally across their backs while the females stay drab. Aud the true purpose of these orange bars is unknown though they bring up questions. These lizards also pair bond so they will have a mate when they come out of hibernation for the mating season. Because of this, females get to choose which males they want to mate and pair bond with based on their preference of phenotypic traits of the males.
Much of this helped us bring up the question that we are asking which is "Do either female collared lizard hatchlings and!or yearlings prefer males with bright orange bars over the males that are drab?" Understanding this question will help with understanding the questions behind the mystery of the oranges bars. I hypothesize that females would have a greater preference towards the Hatchling orange barred males which will be a delayed benefit towards fitness. I hypothesized this because if the females preferred males with orange bars then there would be less of an availability of mating with males making for a smaller amount of clutches leading to a delayed benefit towards the lizard's fitness.
In order to try to prove my hypothesis for this project we will go out to our site at sooner lake; we have sites on both the dam known as SL! and in an area along the shoreline known as SL2. By use of the technique known as noosing we will capture about 20+ of both males and females ranging from hatchlings to yearlings so as for a larger group of test subject. The lizards will be transported back to our lab at the Stillwater campus. We will use three tanks for the lizards during trials, two tanks will be set up identical to each other side by side, horizontally, but have the view of both tanks side blocked from sight so the males will not be able to see each other.
We will observe the decisions of the females over a period of one to two hours to see which male she prefers. Her preference will be determined based on which of the two males she will stay closest to or further from the longest. I expect that the females will choose the females will choose the males with these orange bars over their drab counterparts. Though I do expect that there is a possibility that my hypothesis is wrong and the females might choose the drab males over the vivid males or they may choose neither.
Sharon Simon, Biochemistry

Research: Contagious Laughter in Babies
[View my work!]
Project Summary: Empathy is a high emotional state that provides individuals or groups a stronger social connection. The process of empathy can be observed to be one of the many factors of moral development of the human mind, and can lead to positive outcomes in most social situations or relationships. One of the questions of this particular moral behavior that leads to this research is, "When exactly does empathy begin to develop (at what age in particular) and is there a specific event that triggers empathy-related neural impulses in the brain?" Previous studies show how the contagious crying phenomenon plays a strong effect amongst infants and is one of the earliest stages of moral development. Seeing or hearing emotional distress from an infant can create a chain reaction with infants nearby, and cause them to produce signs of distress through facial expressions, body language, references to mothers, and ultimately vocal responses, supporting the concept of emotional contagion.
There are many theories as to why this occurs. An idea in particular behind why humans emotionally feel the same as another is based on the concept of "mirror neurons." Mirror neurons are neurons in the brain that assist the individual observing the nearby individual to visualize themselves in the same actions as the other. There have also been more studies on the neurological aspect of empathy that indicates that the region of the brain where empathy is active is the same region where neural impulses of physical pain are active.
This idea is an example of why humans react with strong facial distress and upper body movement such as cringing when another person is seen getting physical injured. Another question in the research is, "Can this effect occur in infants with positive emotional states such as laughing in correlation to the development of empathy?" Could there be a contagious laughing phenomenon? The lab's approach to achieving answers to these questions begins by trying to gain as many participants as possible. The approach to finding infants for the study is tedious and acquires an inconsistent rate in finding new participants.
The procedures include creating and distributing fliers that clearly advertise the purpose behind the research and state the compensation, putting the study in local newspapers, and approaching strangers or known people. Only mothers with 5-month-old infants are asked to participate in the study since the act of laughter does not occur until infants reach three to four months. The infants are also brought back at 10 months for an update on their emotional responses. In order to get a clearer understanding of the psychological state of the infants during the experiment, the study records the physiological aspect of the infant in order to clarify the correlation between mind and body. By testing the heart rate and the cortisol levels, the state of the infant will made clearer. The study is still being conducted and no final results have been gathered. The process of finding infants, performing the experiment, and coding the recorded videos are still ongoing. The concept of contagious laughing is hypothesized by the lab that there can be a positive emotional contagion, and the act of infants laughing can psychologically encourage the infants nearby to laugh in unison.
Madison Slawson, Biochemistry & Molecular Biology

[View my work!]
Project Summary: In the Ruhl Lab, our research is focusing on the ten proteins that make up the SWI/SNF complex in order to determine which protein functions directly with INI-1. The SWI/SNF complex is also known as the chromatin-remodeling machine. The SWI/SNF complex plays a key role in the regulation of eukaryotic gene expression. Each of the ten proteins plays an important part in the complex, due to their strong correlation to several cancer types. We currently are focusing on the INI-1 protein of the complex, which has shown to be lacking in atypical teratoid/ rhabdoid tumors (AT/RT). AT/RT is a highly malignant central nervous system tumor often misdiagnosed as some other type of pediatric embryonal tumor. Genetic studies have shown that deletion or mutation of the INI1 gene, which is located on 22ql 1.2, occurs in AT/RT lesions. In our lab, we are trying to understand the biochemistry of the tumor suppressor INI-1 and its role within the SWI/SNF complex. With regard to AR/RT, we are trying to determine the identifying characteristics of INI-1 in the complex, if some patients are missing a portion of or the entire amino acid sequence. If we are able to determine the function ofINI-1 in the SWI/SNF complex, we will be able to determine how to treat patients with this condition.
We have determined that INI-1 is missing from the DNA structure in children who develop AR/RT at such a young age. Using polyacrylamide gel without SDS (native gel), we are investigating the shape and size of the protein. After determining the characteristics of INI-1, we will be able to move further into research of INI-1. We believe that this research will provide evidence that this type of juvenile cancer is present in patients who lack partial protein strands, as well as those lacking entire protein strands, allowing doctors to eventually treat patients with AR/RT more successfully.
Based on the results of this study, our research could provide information that will allow pharmaceutical scientists to develop drug treatments specific to incomplete or missing protein strands. In turn, medical doctors will be able to prescribe a more specific treatment plan for AR/RT patients based on this information. Currently, doctors are using the same treatment for all patients with this condition. At this time, there is only a small chance of curing patients with AR/RT using available treatment options. Pharmaceutical scientists and medical doctors need additional information about the missing or partially missing protein strands known to be associated with this type of cancer in order to develop successful treatment therapies for patients. Our research will be able to provide the medical community with a better understanding of the cause and treatment of this condition.
Sydni Smith, Microbiology & Molecular Genetics
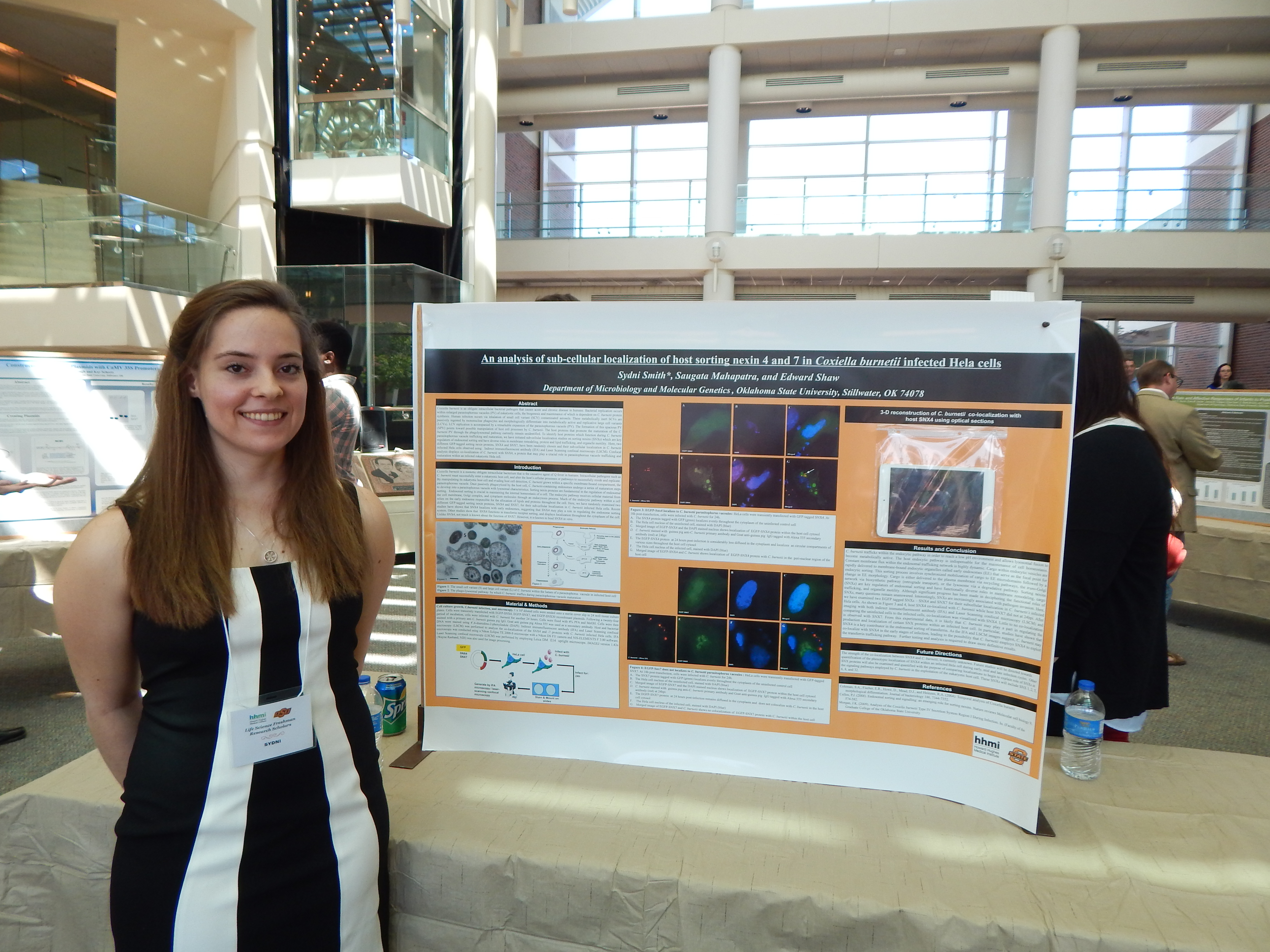
Research: The Utilization of Epigenetics to Further Understanding of Molecular Interaction Pathways Between Coxiella burnetii and its Eukaryotic Host Cell
[View my work!]
Project Summary: Coxiella burnetii is a zoonotic obligate intracellular bacterium that is the causative agent of Q fever in humans; it harbors the distinct potential for usage as a biological warfare agent due to its low infectious dose and ability to survive without a host for long periods of time [1]. Being resistant to heat, pressure, and chemical stress, C. burnetii has the ability to survive for long periods of time in nearly all environments [4]. The main mode of infection of C. burnetii is aerosol transmission, with sexual, person-to-person, and tick-borne transmission being rare [4].
Intracellular pathogens such as C. burnetii must be able to successfully enter a eukaryotic host cell, and alter the cellular processes or pathways in order to replicate within the cell. [6]. By manipulating its eukaryotic host cell and evading host cell detection, C. burnetii is able to replicate within a specific niche, the parasitophorous vacuole in its eukaryotic host cell [3]. C. burnetii infects its host by passively allowing phagocytosis, whereby the host cells internalizes the bacterium ; once inside the host, C. burnetii begins to manipulate its vacuole niche to allow for replication; the exact
mechanisms by which it accomplishes this are unknown [5].
This ability of bacterial pathogens such as C. burnelii to modulate their host cell's signaling pathways has become an emerging theme in recent research [1]. Recent research has identified the ability of intracellular bacteria proteins to alter the genetic expression of the host cell, leading to increased fitness of the bacteria [6]. Intracellular bacteria may do this either directly or ind irectly by concerting the gene promoters of a eukaryotic cell through epigenetic modifications [6] . Epigenetic mechanisms within an organism regulate nearly all biological processes from birth to death [2]. Epigenetic deregulation is the cause of many diseases, highlighting its importance in overall health [2]. C. burnetii protein synthesis may modify epigenetic host cell gene expression to allow for survival and replication within the host cell without a host cell immune response [3]. Focused research into this subject is vital to understanding C. burnetii and, by extension, Q fever [3]. Studies addressing the changes in the epigenome of infected cells, whether infection is parasitic or symbiotic, are rare [6].
It is hypothesized that the obligate intracellular bacterium C. burnetii modulates its eukaryotic host cell's gene expression during infection. More specifically, epigenetic analysis may be essential in further understanding the mechanisms with which C. burnetii exploits its host cell. Various commercially available array kits have the ability to "track" epigenetic changes that have occurred in a cell through the use of antibodies.
Two array kits in particular will be utilized in these studies: PCR chromatin enzyme modification and DNA methylation detection. These will be used in tangent to accurately identify any epigenetic shifts in a healthy cell compared to a C. burnetii infected cell, providing evidence of the means by which C. burnetii may manipulate its eukaryotic host cell during infection.
Jack Spicer, Zoology

[View my work!]
Project Summary: Dr. Wouter Hoff has been researching photoactive yellow proteins (PYP) for some time. Since PYP is light reactant it allows us to study the reactions of proteins the instant they occur by exposing them to a high power laser instead of traditional substrates in solutions. Traditional methods could take seconds to minutes whereas proteins can be exposed to a laser in less than a nanosecond. The main question Dr. Hoff is researching is how the shape of these proteins effects the reactions, and if subtle mutations can affect the way the proteins react. To do this he needs a constant source of purified PYP samples along with purified mutant samples to compare them too. This is where my work comes in. My job is to incubate in E. Coli bacteria that have been genetically modified to produce PYP, harvest the proteins after they have been produced and then purify them thoroughly so that they can be tested by Dr. Hoff and his team.
I begin this process by incubating genetically modified E. Coli for 16-20 hours at a temperature of 37 degrees Celsius in LB broth. The antibiotic ampicillin is added during this process to keep any other bacteria from growing in our broth. After the first incubation, a solution called !PIG is added at a ratio I ml per I L of broth. IPTG activates the production of PYP in the incubated E. Coli. The broth is then incubated for another 4-6 hours. After this final incubation stage the samples are put in a centrifuge for I 0 minutes at a speed of four thousand rotations per minute. This spins down all the E. Coli into a solid pellet and allows us to pour out the broth that is resting on top of it. Now that the E. Coli has been activated to produce PYP and has been separated from the broth. I am able to disrupt the E. Coli and release the PYP by suspending the pellet and using a process called sonication. This is when we expose to the samples to incredibly high sound frequencies to destroy the E. Coli's membrane. You expose them for one minute, five times to ensure that the E. Coli is thoroughly disrupted. During this process the samples must be on ice in order to keep the heat produced by the sonication from denaturing the target proteins. After this a very small sample of about 200 microliters is centrifuged. Ifthe pellet at the bottom is still yellow it means that not all of the E. Coli was disrupted and sonication needs to be repeated. Ifthe pellet is not yellow then the samples are centrifuged at a speed of 20k rpms for twenty minutes. This centrifugation spins down all of the broken E. Coli membranes, and allows us to collect the now free floating proteins that were released during sonication, PYP being one of them.
The next step of my work after the PYP is extracted from the E. Coli is purification. This step is much simpler as it involves rinsing the samples through multiple columns that catch PYP and allow other unwanted proteins to wash out with buffer. Between each filtration we check the spectrum absorption of the remaining protein in the sample. Since PYP as a distinct yellow coloring we look for a large absorption rate at a spectrum frequency of about 450 abs. This is how we know we make the sample more pure with each filtration. It is also how we ensure are final product is a pure highly concentrated sample of PYP.
Sierra Tessmann, Biochemistry & Molecular Biology
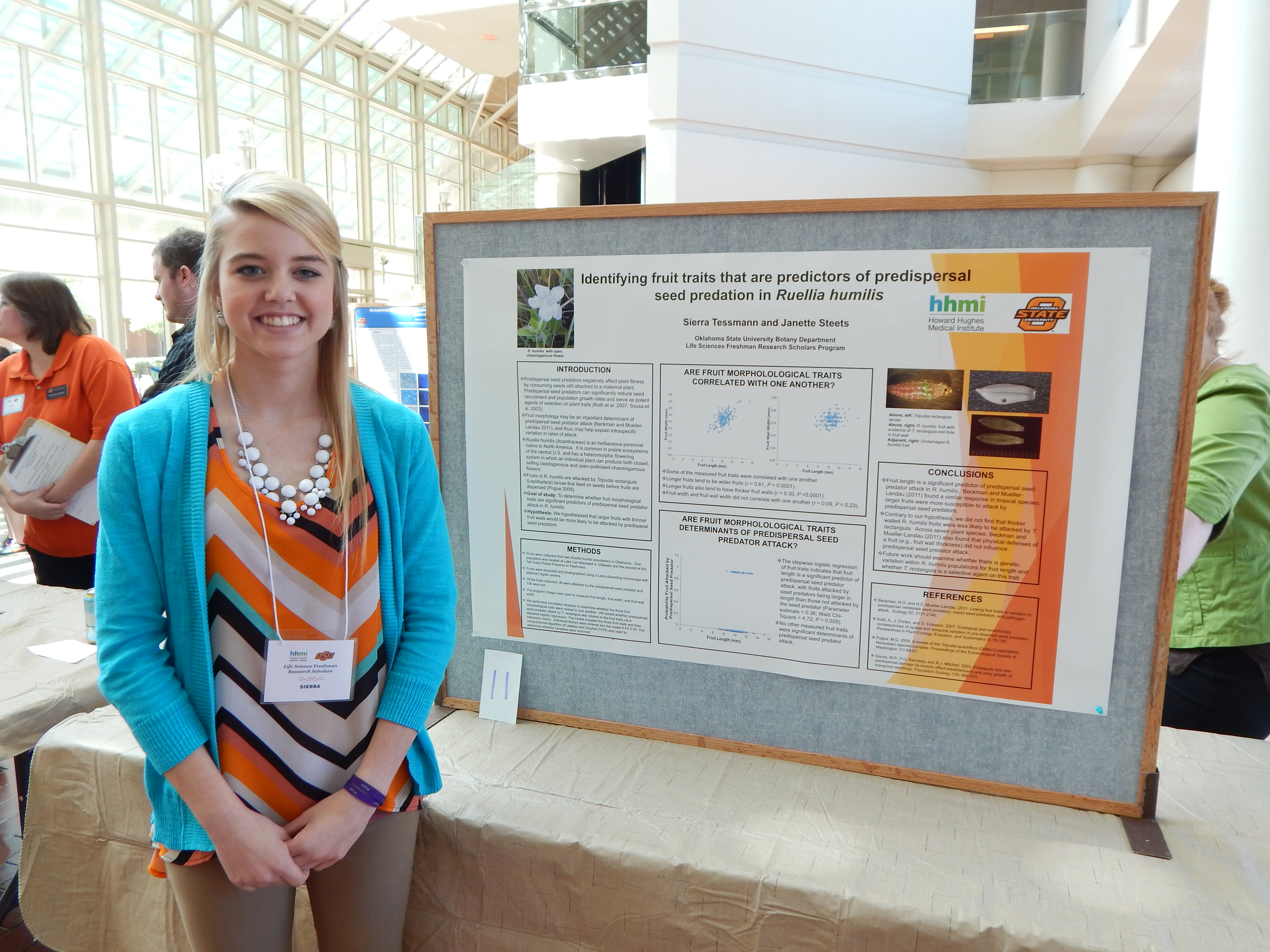
Research: Effects of Seed Predation on Fruits of Ruellia humilis
[View my work!]
Project Summary: Predispersal seeds predators negatively affect plant fitness by consuming seeds still attached to a maternal plant (Kolb et al. 2007). As predispersal seed predators (hereafter, seed predators) can significantly reduce seed recruitment and population growth rates (reviewed in Kolb et al. 2007), it is important to identify morphological traits related to seed predator attack. Seed and fruit morphology may be important in determining interspecific variation in susceptibility to seed predation (Leishman et al. 2000). In some systems, seed size is positively correlated with insect seed predation (Mucunguzi 1995, Espelta et al. 2009). In addition, larger fruits tend to be attacked at a higher rate than smaller fruits (Gomez and Zamora 1994).
This study is important because pre-dispersal seed predation may have important effects on population dynamics and trait evolution in plants. The correlation between seed predators and fruit attack is said to be strong when it affects plant population dynamics and there are changes in the overall plant trait-fitness relationships. The correlation is said to be weak between seed predators and plant species when there is no effects on the overall evolutionary traits of the species (Kolb et al. 2007).
In this study, we will examine whether fruit traits of Ruellia humilis (Acanthaceae) are related to seed predator attack rate. Ruellia humilis is an herbaceous plant native to Native America and is typically found in prairies, open wooded areas, sandy and textured soils, and graveled slopes (Baskin & Baskin, 1982; Long, 1961; Long & Uttal, 1962; USDA-NRCS, 2011). The fruits of Rue /lia humilis are attacked by a specialist seed predator, Tripudia rectangula (Lepidoptera: Noctuidae) (Pogue 2009). T. rectangula larvae feed on the seeds within a fruit before the fruits are dispersed from the maternal plant.
Individual plants of Rue Ilia humilis produce two different types of flowers: open, showy chasmogamous (CH) flowers that are open to pollination by insect vectors and closed, self fertilizing clcistogamous (CL) flowers. In this study we will determine whether CH and CL fruit length and width are related to T. rectangula attack rate. We hypothesize that the seed predator will have a higher attack rate oflarge than small fruits. Fruits were collected from two R.
humilis populations: Lake Carl Blackwell, Stillwater, OK and Tall Grass Prairie Preserve, Pawhuska, OK. Fruits will be inspected and categorized as damaged or undamaged by T. rectangula. Then fruits will be photographed using a Leica dissecting microscope and camera system. Fruit length and width will be measured using image analysis software (Image J). We will use statistical analyses to determine whether R. humilis fruit traits relate to T. rectangula attack.
Kendall Wilson-Henson, Zoology

Research: The Pros and Cons of Prepubescent Coloration
[View my work!]
Project Summary: I am currently studying a species of lizards called Eastern Collared Lizard (Crotaphytus collaris collaris). I investigate the abnormal prepubescent coloration of male hatchlings. my main question is whether these orange bars relate to the overall fitness of the males. my study explores the pros and cons associated with hatchlings having this bars based on how predators react to these bars. Since the bars stand out against the lizards natural habitat I assume this will lead to a heavier predation rate by Coachwhip snakes (Masticophis flagellum). My hypothesis is that Coachwhips will predate the male hatchlings with vivid orange bars more then drab colored hatchlings.

Figure 1: aboveThe Black arrow points to the black collar that gives these lizards their name. The orange arrow points towards the prepubescent coloration that is predominant in male hatchlings. This means males from time of hatch till their second summer.
These bars are important, because they show themselves before the hatchling reach an age of puberty. Their appearance, before puberty, is highly confusing since animals should not display sexual dimorphism during non-sexual years. Dimorphism is the occurrence of gender specific formations found in one species.
I will be monitoring two populations that are located on the outside edges of Stillwater, Ok. I will be taking male hatchlings pairs from both populations for separate trials. These pairs will be places in separate tanks that will allow us to monitor a Coachwhips reaction to the hatchling pairs. We will be running the trials with the same pair for three days total. Once the trails are over the pairs will be placed back into the population in the same locations where they were originally found. These trials will be repeated multiple times until I have gained conclusive data from them.
The trials will be conducted continuously over the three day period. Since a person can not be present continuously the trials will be conducted in the presence of a game video recorder. This will allow me to have permanent records of the trials for complete analysis. This approach has been commonly utilized in animal behavior studies. It allows me to monitor all animals with out any bodily harm coming to any of the animals. The ways my trials could fail is if the Coachwhip does not show interest in either subjects. This might be caused by the smell of paint on the lizards, or could be due to over feeding of the snake. Both of these cases are caused by human error. However, If the Coachwhip show higher interest and predation reaction to the drab hatchlings this would indicated that my original hypothesis is incorrect. I would then need to go back and re- evaluate the hatchling coloration hypothesis. My expectation is that the Coachwhip will focus on the vividly colored hatchlings. Therefore, we shall see a significant con in the males orange bars.